DNA: Difference between revisions
(Rewrote) |
|||
| Line 1: | Line 1: | ||
[[File:3llaa8b7olsb1.jpg|thumb|right|Initial QC’d DNA Samples]] | [[File:3llaa8b7olsb1.jpg|thumb|right|Initial QC’d DNA Samples]] | ||
=== Abstract === | |||
The genomic analysis of three mummified remains from Nazca, Peru, sparked interest due to speculations about extraterrestrial origins. DNA sequencing revealed significant contamination and degradation consistent with ancient remains. The study found no evidence of extraterrestrial or genetically engineered origins but identified various microbial DNA and traces of human DNA, raising questions about their origins and genetic history. | |||
=== Introduction === | === Introduction === | ||
In | In the Nazca region of Peru, several tridactyl mummies were discovered between 2016 and 2017, sparking debates about their origins, with some speculating extraterrestrial links. The Universidad Nacional San Luis Gonzaga in Peru took custody of these mummies and invited scientific studies. This research seeks to understand the genomic information within these mummies using bioinformatics. | ||
The | |||
=== Methods === | |||
Three samples of ancient DNA were analyzed using Next-Generation Sequencing. Data preparation involved quality control and trimming processes to ensure clean datasets. Taxonomic classification was done using Kraken2 against a comprehensive "nt" database. DNA sequences were analyzed through alignment with the human genome and de novo assembly for non-human parts. The study also included mitochondrial DNA analysis to trace lineage. | |||
=== Results === | |||
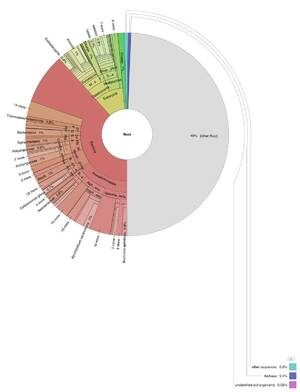
The results confirmed the presence of ancient, degraded DNA heavily influenced by microbial life. A significant finding was the strong alignment of one sample's mitochondrial DNA with the M20a haplogroup, indicating a link to a Southeast Asian lineage, diverging from the expected indigenous American lineage. | |||
=== Discussion === | |||
The findings reveal a complex genetic picture with no evidence of extraterrestrial DNA. The presence of microbial DNA is consistent with ancient samples, while the unexpected Southeast Asian genetic link in one sample invites further questions about human migration and ancient intercontinental connections. | |||
=== | === Conclusion === | ||
This research contributes to understanding the genetic makeup of ancient Peruvian mummies. The lack of extraterrestrial DNA reaffirms the terrestrial nature of these remains and opens avenues for further exploration of human history and migration patterns. | |||
=== Supplemental Materials === | |||
* [https://verbalcant.github.io/nazca_mummies.html Project Homepage: Nazca Mummies Research] | |||
=== | * [https://github.com/VerbalCant/peru_mummy_pipeline GitHub Repository: Peru Mummy Pipeline] | ||
* [https://usegalaxy.org/u/verbal_cant/h/perumummyphase1 Data Files: Galaxy Project] | |||
* [https://www.ncbi.nlm.nih.gov/bioproject/PRJNA865375 Ancient0003 (Maria)] | * [https://www.ncbi.nlm.nih.gov/bioproject/PRJNA865375 Ancient0003 (Maria)] | ||
* [https://www.ncbi.nlm.nih.gov/sra?linkname=bioproject_sra_all&from_uid=869134 Ancient0002 (Victoria)] | * [https://www.ncbi.nlm.nih.gov/sra?linkname=bioproject_sra_all&from_uid=869134 Ancient0002 (Victoria)] | ||
| Line 30: | Line 29: | ||
* [https://www.bioinformaticscro.com/blog/dna-evidence-for-alien-nazca-mummies-lacking/ The Bioinformatics CRO] | * [https://www.bioinformaticscro.com/blog/dna-evidence-for-alien-nazca-mummies-lacking/ The Bioinformatics CRO] | ||
* [https://www.reddit.com/r/UFOs/comments/17o84r6/mummys_the_word_a_genomic_look_at_peruvian_mummies/?share_id=hKM-AEWJUTijpAhKi731I Bioinformatics Expert Analysis] | * [https://www.reddit.com/r/UFOs/comments/17o84r6/mummys_the_word_a_genomic_look_at_peruvian_mummies/?share_id=hKM-AEWJUTijpAhKi731I Bioinformatics Expert Analysis] | ||
=== References === | |||
- Comprehensive list of references and sources used in the study, including specific papers, databases, and bioinformatics tools. | |||
{{#seo:|title=DNA Analysis}} | {{#seo:|title=DNA Analysis}} | ||
Revision as of 01:48, 16 March 2024
Abstract
The genomic analysis of three mummified remains from Nazca, Peru, sparked interest due to speculations about extraterrestrial origins. DNA sequencing revealed significant contamination and degradation consistent with ancient remains. The study found no evidence of extraterrestrial or genetically engineered origins but identified various microbial DNA and traces of human DNA, raising questions about their origins and genetic history.
Introduction
In the Nazca region of Peru, several tridactyl mummies were discovered between 2016 and 2017, sparking debates about their origins, with some speculating extraterrestrial links. The Universidad Nacional San Luis Gonzaga in Peru took custody of these mummies and invited scientific studies. This research seeks to understand the genomic information within these mummies using bioinformatics.
Methods
Three samples of ancient DNA were analyzed using Next-Generation Sequencing. Data preparation involved quality control and trimming processes to ensure clean datasets. Taxonomic classification was done using Kraken2 against a comprehensive "nt" database. DNA sequences were analyzed through alignment with the human genome and de novo assembly for non-human parts. The study also included mitochondrial DNA analysis to trace lineage.
Results
The results confirmed the presence of ancient, degraded DNA heavily influenced by microbial life. A significant finding was the strong alignment of one sample's mitochondrial DNA with the M20a haplogroup, indicating a link to a Southeast Asian lineage, diverging from the expected indigenous American lineage.
Discussion
The findings reveal a complex genetic picture with no evidence of extraterrestrial DNA. The presence of microbial DNA is consistent with ancient samples, while the unexpected Southeast Asian genetic link in one sample invites further questions about human migration and ancient intercontinental connections.
Conclusion
This research contributes to understanding the genetic makeup of ancient Peruvian mummies. The lack of extraterrestrial DNA reaffirms the terrestrial nature of these remains and opens avenues for further exploration of human history and migration patterns.
Supplemental Materials
- Project Homepage: Nazca Mummies Research
- GitHub Repository: Peru Mummy Pipeline
- Data Files: Galaxy Project
- Ancient0003 (Maria)
- Ancient0002 (Victoria)
- Ancient0004 (Victoria)
- Abraxas Biosystems Report
- Genomic Analysis
- The Bioinformatics CRO
- Bioinformatics Expert Analysis
References
- Comprehensive list of references and sources used in the study, including specific papers, databases, and bioinformatics tools.